Modulating Autophagy as an Attractive Strategy to Treat Renal Cell Carcinoma
| Received 25 Oct, 2023 |
Accepted 04 Apr, 2024 |
Published 31 Dec, 2024 |
Autophagy is a key major mechanism by which cancer cells are able to survive periods of therapy-induced stress that result in drug resistance, autophagy is crucial for mediating cellular stress and damage. Renal Cell Carcinomas (RCC) make up about 90% of kidney cancer cases. Modulating autophagy has been demonstrated to increase the cytotoxicity of authorized Renal Cell Carcinoma (RCC) therapies and combat drug resistance in recent years. Autophagy activation inhibits malignancies by removing damaged organelles and proteins while shielding certain tumor cells from low oxygen levels and starvation, which are primary features of tumor microenvironments. It is unknown why autophagy has two effects, despite evidence that it controls both cell death and survival. Research has shown that autophagy regulators can treat RCC by controlling the PI3K/Akt/mTOR and AMPK/mTOR signaling pathways. On the other hand, the relationship between autophagy and immune cell activation, especially in RCC, is unclear. Early studies suggest inhibiting autophagy can affect hematopoiesis and systemic immunity, suggesting immune checkpoint inhibitor therapy may not be beneficial. Recent studies show autophagy inhibition does not stop T-cell activation. It is noteworthy that studies examining the connection between autophagy and RCC are becoming more popular. Autophagy-related proteins have been shown in earlier research to be interesting prognostic indicators for Renal Cell Carcinoma (RCC) treatment. Therefore, to determine the therapeutic relevance of autophagy activators and inhibitors in Renal cell carcinomas (RCC), more investigation is required. This study explores the therapeutic alternatives for the current situation and potential future directions of autophagy targeting as a potential alternative and successful treatment for renal cell carcinoma.
INTRODUCTION
Primarily formed from renal tubular epithelial cells, renal cell carcinoma (RCC) is thought to be the most prevalent kind of malignant kidney tumor. The most prevalent subtype of RCC is reportedly renal clear cell carcinoma (ccRCC)1. The biology, pathogenesis and treatment of RCC have advanced, but the incidence of the disease is still rising. More than 200,000 new cases are reported globally each year and their incidence has been rising at a rate of roughly 2% each year2. Even though there are numerous targeted medications and biologicals for ccRCC that are now in the research and development stage, it is still unknown whether they are effective. Nevertheless, numerous research has lately revealed close connections between autophagy and RCC, which may present fresh therapeutic possibilities for RCC3.
|
|
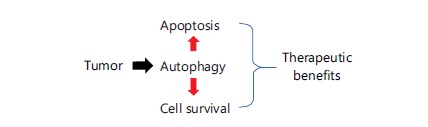
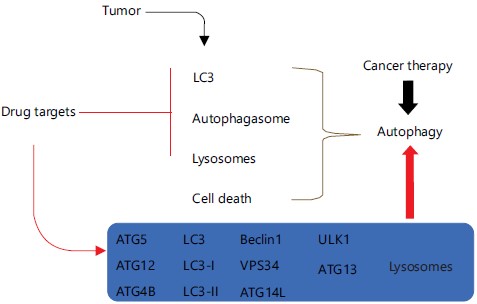
It is important to note that studies on the connection between autophagy and RCC have drawn attention (Fig. 1). Therefore, autophagy-related proteins may serve as promising prognostic indicators for the therapy of RCC, according to prior research (Fig. 2)4-6.
Autophagy is a tightly regulated catabolic system for self-preservation that involves the breakdown of extraneous or malfunctioning parts7. Autophagy-related (ATG) genes play a significant role in controlling this process, which enables cells to adapt to a variety of conditions such as nutrition deprivation, endoplasmic reticulum (ER) stress, pathogen infection, or hypoxia, which is thought to be a survival mechanism8,9. However, autophagy can take part in cell death under severe or protracted stress or in cells lacking in apoptosis10. The ROS is a crucial regulator of both apoptosis and autophagy, however, both processes are accompanied by excessive overproduction, phosphatidylinositol 3-kinase inactivation and mitogen-activated protein kinase (MAPK) activation11-13.
As a result, once autophagy is initiated, it can shield cells from harm caused by aging organelles and damaged macromolecules that can be broken down by lysosomal enzymes. Autophagy, however, appears to be a double-edged sword that can both cause autophagic cell death (type II programmed cell death) and cell survival14 depending on the situation (Fig. 2). The C-terminal peptide of LC3B, a microtubule-associated protein light chain 3, is often the first part of the autophagy process to be broken down by the cysteine protease ATG4 to produce LC3B-I, which is subsequently conjugated to phosphoethanolamine to produce LC3BII. Furthermore, LC3BII can join forces with ATG5, 7 and 12 to create autophagosomes that contain phospholipid bilayers15. In order to degrade macromolecules and damaged organelles, ubiquitin-binding protein p62/sequestosome-1(SQSTM1) interacts with LC3 simultaneously. The AMPK (increased ratio of AMP/ATP, nutrient deficiency) can activate autophagy induction, whereas pmTOR suppresses it. Under conditions of nutrient deficiency, AMPK will be phosphorylated/activated to trigger autophagy initiation, whereas pmTOR, a cell survival signaling, can suppress autophagy formation15. Similarly, these two mechanisms frequently use a variety of paths in response to a single stress. The new target mechanism for chemotherapy-resistant cancer therapy is therefore autophagy/apoptosis16,17.
Depending on the kind, grade, stage and depth of the tumor, autophagy’s effect in cancer can either be tumor-suppressive or tumor-promoting18. Metastatic RCC cells may use autophagy as a means of cell survival and inhibiting it in ccRCC may increase the cytotoxicity of mTOR inhibitors, according to certain studies19,20. When ccRCC patients use pazopanib, autophagic gene polymorphisms are linked to progression-free survival (PFS). Therefore, it is hypothesized that autophagy regulation and function are probably related to the preservation of kidney cancer cell homeostasis, disease etiology and therapeutic resistance.
AUTOPHAGY IN RENAL CELL CARCINOMA
The connection between autophagy and RCC has attracted attention. As a result, investigating the molecular basis of autophagy in RCC may serve as a therapeutic target15. According to research by Radovanovic et al.21, the role of autophagy in ccRCC is independent of AMPK/Mtor transcriptional regulation of autophagy. In ccRCC tumors, the mRNA levels of pro-autophagic ATG4, p62 and UVRAG were higher than those of pro-apoptotic BAX, anti-apoptotic BCL-XL, pro-apoptotic ATG4 and BCL-XL. A rise in phospho-ULK1 and the degradation of the autophagic substrate p62, while leaving apoptotic PARP cleavage unaffected, served as evidence that autophagy had been induced. The AMPK phosphorylation was reduced and 4EBP1 phosphorylation was increased in ccRCC tissue21.
The expression of apoptosis regulators did not correlate with clinicopathological features of ccRCC. On the other hand, higher levels of the mRNA for ATG4, GABARAP and p62 were linked to earlier tumor stages, smaller tumors and improved disease-specific 5-year survival (ATG4 and p62). Therefore, lower tumor stage, less metastasis and improved 5 year survival were related to decreased p62 protein levels, which correspond to greater autophagic flux. In a different study, Deng et al.22 discovered that sinomenine inhibited the PI3K/Akt/mTOR signaling pathway, which caused ACHN cells to undergo cell death and autophagy. Additionally, Antonaci et al.23 demonstrated that PI3K/AKT/mTOR/p70S6K pathways were involved in dimethyl sulfide (DMS)-induced autophagy in Caki-1 cells. Autophagy has been activated in vivo and in vitro by sunitinib inhibiting the Akt/mTOR/p70S6K pathway24. The literature reviewed above demonstrates that autophagy and the PI3K/Akt/mTOR signaling pathway may serve as therapeutic targets for the prevention and treatment of RCC. The basal amount of autophagy is inherently higher in RCC cell lines. One study found that between 30 and 60% of growing cells in different RCC cell lines have LC3-II puncta25.
IMBALANCE IN AUTOPHAGY MARKERS IN RENAL CELL CARCINOMA (RCC)
Beclin1 participates in the formation of Beclin 1-Vps34-Vps15 core complexes and the start of autophagy, especially in unfavorable conditions26. It was discovered that RCC tissues and cell lines (A498 and ACHN) express Beclin 1 at high levels4. Reduced expression of ATGs, which are essential for the process of autophagy nucleation, is associated with a poor prognosis in RCC27,28. According to Liu et al.29, the majority of ccRCCs show allelic loss and/or mutations of ATG7 p62/SQSTM1, a conventional macroautophagy substrate, interacts with LC3 directly to digest ubiquitinated protein30. Autophagy decreased p62 levels in RCC and additionally, p62 amplification on chromosome 5 was linked to the development of kidney carcinoma31.
Studies have shown that the ccRCC, A498 and ACHN cell lines’ up-regulated expression of LC3 encourages cell motility4. According to two investigations, LC3-II levels in RCC tissues and cells were lowered by boosting autophagy-related apoptosis24,32. However, Wang et al.33 discovered that LC3-II expression levels in RCC cell lines (786-O, 769-P, OS-RC-2 and ACHN cells) were lower than those in a control cell line (HK-2 cell).
AUTOPHAGY CONTROLS BOTH CELL DEATH AND SURVIVAL
It is unclear why autophagy has two effects. Numerous investigations have shown that autophagy regulates both cell survival and death (Fig. 2). Reduced and aberrant autophagy gene and protein expression may have an effect on RCC illness. The autophagy-related proteins Beclin-1 and LC3B-II have been demonstrated to be downregulated in kidney cancer cells by the cytoprotective enzyme heme oxygenase-1 (HO-1)33. Wang et al.33 found that Atg7 and LC3-II overexpression suppressed cell proliferation in the human RCC cell lines 786-O, 769-P, OS-RC-2 and ACHN both in vivo and in vitro. However, autophagy protects some tumor cells against nutrient deprivation and low oxygen levels, which are the two main characteristics of tumor microenvironments34. In RCC cells lacking VHL, EPAS1, a type of hypoxia-inducible factor, builds up and targets ITPR1. In the interim, ITPR1 regulates the recognition of an unidentified signal coming from NK cells that initiates autophagy. Granzyme B (GZMB), a substance that NK cells make, is destroyed as a result of autophagy activation in RCC cells, which reduces NK’s capacity to eradicate tumor cells35,36. Furthermore, it has been shown that ITPR1 regulates the induction of autophagy in the NK-mediated killing of RCC cells36.
IMMUNE SYSTEM AND AUTOPHAGY
Despite the fact that it is uncertain how autophagy suppression affects the effectiveness of chemotherapies, clinical trials have started utilizing CQ or HCQ, primarily in conjunction with them, to treat cancer patients37-39. These combinations, which are mostly used to treat cancer in xenograft mouse models, have demonstrated some promise in suppressing tumor growth and extending host survival40-42. However, to prevent tumor rejection in these studies, immuno-deficient animals were used, therefore there was no chance to assess how autophagy inhibitors affect immune system cells directly and indirectly43. Further studies employing immune-competent mice have shown that the loss of essential autophagy-relevantgene products such as autophagy-related (ATG) 5 or Beclin 1 (BECN1) reduces the effectiveness of radiotherapy or chemotherapy in immune-competent mice, even though these treatments have a greater anti-cancer effect in vitro and in vivo in immune-deficient mice44.
The study’s finding highlights the essential role that the immune system plays in effective anticancer treatment when autophagy is being altered. The immune system may be stimulated by immunogenic cell death (ICD), which was demonstrated to be produced by a number of substances released by cancer cells as they died following chemotherapies or when exposed to the environment on their surface45,46. The Damage Associated Molecular Patterns (DAMPs) exposed on the surface of dying lymphoma cells treated with bortezomib were Calreticulin and Heat Shock Protein (HSP) 90 and that the receptor molecule involved in dendritic cells’ (DCs’) recognition of these DAMPs was CD9147,48. This protein was found to be significant in the study. Strong antigen-presenting cells (APCs) called DCs are necessary for inducing a specific immune response and destroying cancer cells that have undergone apoptosis49. Therefore, it is now plainly clear that the immune system’s participation is necessary for effective anticancer therapy.
TARGETING AUTOPHAGY IN RENAL CELL CARCINOMA
Cancer cells can provide alternate energy sources in times of stress by recycling nutrients via autophagy, a critical lysosomal breakdown mechanism50,51. Numerous recent studies have suggested that changes in the autophagy pathway may be particularly significant for patients with RCC and affect overall survival52,53. For the treatment of cancer, some substances pharmacologically target autophagy. Additionally, lucanthone is being researched as a cancer preventative. In tests using cell culture, lucanthone revealed lysosomal disruption and suppression of autophagy53. Furthermore, cathepsin D, a lysosomal protease, was discovered to be an important mediator of lucanthone’s powerful pro-apoptotic actions in a number of breast cancer cell lines. Researchers have developed new, lysosome-targeting medications with the use of insights from the chemical composition of lucanthone. The STF-62247 specific substance is thought to trigger autophagy in cancer cells since it has strong lethal effects on VHL-deficient cancer cells but is ineffective against wild-type (WT) VHL cells. Treatment results in huge, cytoplasmic vacuoles in both WT-VHL and VHL-deficient cells. The molecular connections between VHL and autophagy, however, have not yet been thoroughly clarified53.
Both chloroquine (CQ) and hydroxychloroquine (HCQ) substances function by building up in lysosomes and then deacidifying them54. This deacidification prevents autophagy from taking place because lysosomes’ low pH is necessary for the cargo to degrade. The CQ and HCQ have been repurposed to pharmacologically target autophagy in a range of cancer types for more than 10 years55. However, there haven’t been many clinical investigations that have looked at HCQ in RCC patients. The HCQ and lucanthone core motifs were designed into the dimeric compound ROC-325. Similar to HCQ, ROC-325 concentrates on the late stages of autophagy. Instead of impairing the growth of autophagosomes, ROC-325 assembles in the lysosome and deacidifies it. The indicators LC3-II and p62 stabilize when ROC-325 is applied in vitro, which is consistent with autophagy suppression. At half maximum inhibitory concentrations (IC50) of 2-10 vs. 50-100 M, ROC-325 dramatically decreased cell viability in RCC cell lines as compared to HCQ. Another study found that the suppression of autophagy with 3-methyladenine (3-MA) and bafilomycin dramatically boosted paclitaxel-activated apoptosis in FLCN-deficient RCC cells56,57.
Interestingly, bafilomycin A1 was found to disrupt autophagy by inhibiting the union of the autophagosome and lysosome. These results imply that while these drugs partially inhibit the lysosomal degradation of cellular components, they might not be potent enough to completely inhibit the degradation of autophagy. It is essential to create new, more potent autophagy inhibitors in order to increase the efficacy of treatments.
CONCLUSION AND FUTURE PERSPECTIVE
Autophagy has recently been found to be the main mechanism by which cancer cells can endure periods of therapy-induced stress that lead to drug resistance. The progression of clear cell renal cell carcinoma (ccRCC) continues to be a significant clinical issue and understanding the molecular triggers of malignancy progression is necessary to creating effective therapy targets. The connection between autophagy and immune cell activation, particularly in RCC, is poorly understood. However, preliminary outcomes from a number of cancer models have given conflicting outcomes. Early studies indicate that inhibiting autophagy can have an impact on hematopoiesis and systemic immunity, indicating that using immune checkpoint inhibitor therapy in conjunction with autophagy may not be beneficial. However, recent studies indicate that inhibiting autophagy does not stop T-cell activation. This underlines the requirement for developing and evaluating medicines that modulate autophagy in addition to the expanding list of approved RCC therapies.
SIGNIFICANCE STATEMENT
Renal Cell Carcinoma accounts for malignant renal tumors and its occurrence has been rising at a pace of roughly 2% per year. Consequently, it is essential to create new targets for RCC. However, several recent research has revealed close connections between autophagy and RCC, which may present new avenues for the disease’s treatment. Autophagy inhibits the growth of tumors by removing oxidative stress, preserving genomic stability and lowering malfunctioning proteins in RCC. While, RCC-specific chemotherapy stimulates autophagy, which results in medication tolerance and accelerates the growth of the tumor. This suggests that medications that inhibit autophagy may be useful in the treatment of RCC. In other words, especially for RCC that is resistant to chemotherapy, chemotherapies in combination with autophagy inhibitors may prove more efficacious.
REFERENCES
- Gan, C.L. and D.Y.C. Heng, 2020. New insights into the obesity paradox in renal cell carcinoma. Nat. Rev. Nephrol., 16: 253-254.
- Siegel, R.L., K.D. Miller and A. Jemal, 2019. Cancer statistics, 2019. CA: Cancer J. Clin., 69: 7-34.
- Mikami, S., M. Oya, R. Mizuno, T. Kosaka, K.I. Katsube and Y. Okada, 2014. Invasion and metastasis of renal cell carcinoma. Med. Mol. Morphol., 47: 63-67.
- Guo, Y., H.C. Zhang, S. Xue and J.H. Zheng, 2019. Receptors for advanced glycation end products is associated with autophagy in the clear cell renal cell carcinoma. J. Cancer Res. Ther., 15: 317-323.
- Brodaczewska, K.K., C. Szczylik, M. Fiedorowicz, C. Porta and A.M. Czarnecka, 2016. Choosing the right cell line for renal cell cancer research. Mol. Cancer, 15.
- Asakura, T., A. Imai, N. Ohkubo-Uraoka, M. Kuroda and Y. Iidaka et al., 2005. Relationship between expression of drug-resistance factors and drug sensitivity in normal human renal proximal tubular epithelial cells in comparison with renal cell carcinoma. Oncol. Rep., 14: 601-607.
- Yang, Z. and D.J. Klionsky, 2010. Eaten alive: A history of macroautophagy. Nat. Cell Biol., 12: 814-822.
- Glick, D., S. Barth and K.F. Macleod, 2010. Autophagy: Cellular and molecular mechanisms. J. Pathol., 221: 3-12.
- Reshi, L., J.L. Wu, H.V. Wang and J.R. Hong, 2016. Aquatic viruses induce host cell death pathways and its application. Virus Res., 211: 133-144.
- Nah, J., D. Zablocki and J. Sadoshima, 2022. The role of autophagic cell death in cardiac disease. J. Mol. Cell. Cardiol., 173: 16-24.
- Pan, S., Y. Qin, Z. Zhou, Z. He and X. Zhang et al., 2015. Plumbagin induces G2/M arrest, apoptosis, and autophagy via p38 MAPK- and PI3K/Akt/mTOR-mediated pathways in human tongue squamous cell carcinoma cells. Drug Des. Dev. Ther., 9: 1601-1626.
- Reshi, M.L., Y.C. Su and J.R. Hong, 2014. RNA viruses: ROS-mediated cell death. Int. J. Cell Biol., 2014.
- Reshi, M.L. and J.R. Hong, 2016. Role of Oxidative Stress in RNA Virus-Induced Cell Death. In: Reactive Oxygen Species in Biology and Human Health, Ahmad, S.I., (Ed.), CRC Press, Boca Raton, Florida, ISBN: 9781032339979.
- Niu, N., Z. Wang, S. Pan, H. Ding and G. Au et al., 2015. Pro-apoptotic and pro-autophagic effects of the Aurora kinase A inhibitor alisertib (MLN8237) on human osteosarcoma U-2 OS and MG-63 cells through the activation of mitochondria-mediated pathway and inhibition of p38 MAPK/PI3K/Akt/mTOR signaling pathway. Drug Des. Dev. Ther., 9: 1555-1584.
- Sui, X., R. Chen, Z. Wang, Z. Huang and N. Kong et al., 2013. Autophagy and chemotherapy resistance: A promising therapeutic target for cancer treatment. Cell Death Dis., 4.
- Reshi, L., H.C. Wu, J.L. Wu, H.V. Wang and J.R. Hong, 2016. GSIV serine/threonine kinase can induce apoptotic cell death via p53 and pro-apoptotic gene bax upregulation in fish cells. Apoptosis, 21: 443-458.
- Levy, J.M.M. and A. Thorburn, 2020. Autophagy in cancer: Moving from understanding mechanism to improving therapy responses in patients. Cell Death Differ., 27: 843-857.
- Jonasch, E., P.A. Futreal, I.J. Davis, S.T. Bailey and W.Y. Kim et al., 2012. State of the science: An update on renal cell carcinoma. Mol. Cancer Res., 10: 859-880.
- White, E., 2012. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer, 12: 401-410.
- Santoni, M., F. Piva, U. de Giorgi, A. Mosca and U. Basso et al., 2018. Autophagic gene polymorphisms in liquid biopsies and outcome of patients with metastatic clear cell renal cell carcinoma. Anticancer Res., 38: 5773-5782.
- Radovanovic, M., S. Vidicevic, J. Tasic, N. Tomonjic and Z. Stanojevic et al., 2020. Role of AMPK/mTOR-independent autophagy in clear cell renal cell carcinoma. J. Invest. Med., 68: 1386-1393.
- Deng, F., Y.X. Ma, L. Liang, P. Zhang and J. Feng, 2018. The pro-apoptosis effect of sinomenine in renal carcinoma via inducing autophagy through inactivating PI3K/AKT/mTOR pathway. Biomed. Pharmacother., 97: 1269-1274.
- Antonaci, G., L.G. Cossa, A. Muscella, C. Vetrugno, S.A. De Pascali, F.P. Fanizzi and S. Marsigliante, 2019. [Pt(O,O'-acac)(γ-acac)(DMS)] induces autophagy in Caki-1 renal cancer cells. Biomolecules, 9.
- Li, M.L., Y.Z. Xu, W.J. Lu, Y.H. Li and S.S. Tan et al., 2018. Chloroquine potentiates the anticancer effect of sunitinib on renal cell carcinoma by inhibiting autophagy and inducing apoptosis. Oncol. Lett., 15: 2839-2846.
- Bray, K., R. Mathew, A. Lau, J.J. Kamphorst and J. Fan et al., 2012. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS ONE, 7.
- Kang, R., H.J. Zeh, M.T. Lotze and D. Tang, 2011. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ., 18: 571-580.
- Liu, X.D., J. Yao, D.N. Tripathi, Z. Ding and Y. Xu et al., 2015. Autophagy mediates HIF2α degradation and suppresses renal tumorigenesis. Oncogene, 34: 2450-2460.
- Deng, Q., L. Liang, Q. Liu, W. Duan, Y. Jiang and L. Zhang, 2018. Autophagy is a major mechanism for the dual effects of curcumin on renal cell carcinoma cells. Eur. J. Pharmacol., 826: 24-30.
- Liu, X.D., H. Zhu, A. DePavia and E. Jonasch, 2015. Dysregulation of HIF2α and autophagy in renal cell carcinoma. Mol. Cell. Oncol., 2.
- Pankiv, S., T.H. Clausen, T. Lamark, A. Brech and J.A. Bruun et al., 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem., 282: 24131-24145.
- Mathew, R., C.M. Karp, B. Beaudoin, N. Vuong and G. Chen et al., 2009. Autophagy suppresses tumorigenesis through elimination of p62. Cell, 137: 1062-1075.
- Grimaldi, A., D. Santini, S. Zappavigna, A. Lombardi and G. Misso et al., 2015. Antagonistic effects of chloroquine on autophagy occurrence potentiate the anticancer effects of everolimus on renal cancer cells. Cancer Biol. Ther., 16: 567-579.
- Wang, Z.L., Q. Deng, T. Chong and Z.M. Wang, 2018. Autophagy suppresses the proliferation of renal carcinoma cell. Eur. Rev. Med. Pharmacol. Sci., 22: 343-350.
- Banerjee, P., A. Basu, B. Wegiel, L.E. Otterbein and K. Mizumura et al., 2012. Heme oxygenase-1 promotes survival of renal cancer cells through modulation of apoptosis- and autophagy-regulating molecules. J. Biol. Chem., 287: 32113-32123.
- Katheder, N.S., R. Khezri, F. O’Farrell, S.W. Schultz and A. Jain et al., 2017. Microenvironmental autophagy promotes tumour growth. Nature, 541: 417-420.
- Messai, Y., M.Z. Noman, M. Hasmim, B. Janji and A. Tittarelli et al., 2014. ITPR1 protects renal cancer cells against natural killer cells by inducing autophagy. Cancer Res., 74: 6820-6832.
- Reshi, L., H.V. Wang, C.F. Hui, Y.C. Su and J.R. Hong, 2017. Anti-apoptotic genes Bcl-2 and Bcl-xL overexpression can block iridovirus serine/threonine kinase-induced Bax/mitochondria-mediated cell death in GF-1 cells. Fish Shellfish Immunol., 61: 120-129.
- Marinković, M., M. Šprung, M. Buljubašić and I. Novak, 2018. Autophagy modulation in cancer: Current knowledge on action and therapy. Oxid. Med. Cell. Longevity, 2018.
- Galluzzi, L., J.M.B.S. Pedro, S. Demaria, S.C. Formenti and G. Kroemer, 2017. Activating autophagy to potentiate immunogenic chemotherapy and radiation therapy. Nat. Rev. Clin. Oncol., 14: 247-258.
- Chude, C.I. and R.K. Amaravadi, 2017. Targeting autophagy in cancer: Update on clinical trials and novel inhibitors. Int. J. Mol. Sci., 18.
- Jia, L., J. Wang, T. Wu, J. Wu, J. Ling and B. Cheng, 2017. In vitro and in vivo antitumor effects of chloroquine on oral squamous cell carcinoma. Mol. Med. Rep., 16: 5779-5786.
- Zhao, X.G., R.J. Sun, X.Y. Yang, D.Y. Liu, D.P. Lei, T. Jin and X.L. Pan, 2015. Chloroquine-enhanced efficacy of cisplatin in the treatment of hypopharyngeal carcinoma in xenograft mice. PLoS ONE, 10.
- Chou, H.L., Y.H. Lin, W. Liu, C.Y. Wu and R.N. Li et al., 2019. Combination therapy of chloroquine and C2-ceramide enhances cytotoxicity in lung cancer H460 and H1299 cells. Cancers, 11.
- Jiang, G.M., Y. Tan, H. Wang, L. Peng and H.T. Chen et al., 2019. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer, 18.
- Michaud, M., I. Martins, Abdul Qader Sukkurwala, S. Adjemian and Y. Ma et al., 2011. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science, 334: 1573-1577.
- Cirone, M., L.D. Renzo, L.V. Lotti, V. Conte and P. Trivedi et al., 2012. Activation of dendritic cells by tumor cell death. OncoImmunology, 1: 1218-1219.
- Kepp, O., L. Senovilla, I. Vitale, E. Vacchelli and S. Adjemian et al., 2014. Consensus guidelines for the detection of immunogenic cell death. Oncoimmunology.
- Cirone, M., L.D. Renzo, L.V. Lotti, V. Conte and P. Trivedi et al., 2012. Primary effusion lymphoma cell death induced by bortezomib and AG 490 activates dendritic cells through CD91. PLoS ONE, 7.
- Garufi, A., G. Pistritto, C. Ceci, L. Di Renzo and R. Santarelli et al., 2012. Targeting COX-2/PGE2 pathway in HIPK2 knockdown cancer cells: Impact on dendritic cell maturation. PLoS ONE, 7.
- Mellman, I., 2013. Dendritic cells: Master regulators of the immune response. Cancer Immunol Res., 1: 145-149.
- Rubinsztein, D.C., P. Codogno and B. Levine, 2012. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discovery, 11: 709-730.
- Yang, Z. and D.J. Klionsky, 2010. Mammalian autophagy: Core molecular machinery and signaling regulation. Curr. Opin. Cell Biol., 22: 124-131.
- Bouhamdani, N., D. Comeau, K. Cormier and S. Turcotte, 2019. STF-62247 accumulates in lysosomes and blocks late stages of autophagy to selectively target von Hippel-Lindau-inactivated cells. Am. J. Physiol. Cell Physiol., 316: C605-C620.
- Turcotte, S., D.A. Chan, P.D. Sutphin, M.P. Hay, W.A. Denny and A.J. Giaccia, 2008. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell, 14: 90-102.
- Homewood, C.A., D.C. Warhurst, W. Peters and V.C. Baggaley, 1972. Lysosomes, pH and the anti-malarial action of chloroquine. Nature, 235: 50-52.
- Amaravadi, R.K., D. Yu, J.J. Lum, T. Bui and M.A. Christophorou et al., 2007. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest., 117: 326-336.
- Zhang, Q., S. Si, S. Schoen, J. Chen, X.B. Jin and G. Wu, 2013. Suppression of autophagy enhances preferential toxicity of paclitaxel to folliculin-deficient renal cancer cells. J. Exp. Clin. Cancer Res., 32.
How to Cite this paper?
APA-7 Style
Reshi,
L., Juen,
S. (2024). Modulating Autophagy as an Attractive Strategy to Treat Renal Cell Carcinoma. Asian Science Bulletin, 2(4), 290-297. https://doi.org/10.3923/asb.2024.290.297
ACS Style
Reshi,
L.; Juen,
S. Modulating Autophagy as an Attractive Strategy to Treat Renal Cell Carcinoma. Asian Sci. Bul 2024, 2, 290-297. https://doi.org/10.3923/asb.2024.290.297
AMA Style
Reshi
L, Juen
S. Modulating Autophagy as an Attractive Strategy to Treat Renal Cell Carcinoma. Asian Science Bulletin. 2024; 2(4): 290-297. https://doi.org/10.3923/asb.2024.290.297
Chicago/Turabian Style
Reshi, Lateef, and Shu-Hui Juen.
2024. "Modulating Autophagy as an Attractive Strategy to Treat Renal Cell Carcinoma" Asian Science Bulletin 2, no. 4: 290-297. https://doi.org/10.3923/asb.2024.290.297

This work is licensed under a Creative Commons Attribution 4.0 International License.




