Cloning, Optimized Expression and Purification of Mycobacterium tuberculosis RpfE Protein in Escherichia coli
| Received 11 Apr, 2023 |
Accepted 15 Apr, 2025 |
Published 30 Jun, 2025 |
Background and Objective: One of the five resuscitation promoting factors (Rpf) encoded by Mycobacterium tuberculosis is RpfE which is antigenic, immunogenic, and probably secretory. This indicates RpfE holds promise as a potential vaccine candidate against tuberculosis. The aim of this study was to clone RfpE and subsequently optimize the expression of mature RpfE protein in Escherichia coli for purification. Materials and Methods: The coding sequence of RpfE was chemically synthesized. Full-length RpfE and a fragment of the gene lacking the signal peptide sequence (encoding mature RpfE) were respectively cloned in pET-23a (+) and pET-28a (+) plasmids. The polyhistidine (His)-tagged recombinant proteins were expressed in E. coli BL21 (DE3) and their cellular localization was determined. The optimal induction conditions were determined for mature RpfE. The protein was purified using a His-tag purification kit, was resolved on SDS-PAGE gel, and detected by western blot using anti-His antibody and serum from a consented tuberculosis patient. Results: Full-length RpfE (~26 kDa) was secreted into broth medium while mature RpfE (multimerized as a band of ~35 kDa) was accumulated in the cytoplasm of E. coli. The optimal expression of the mature RpfE was obtained in Luria-Bertani broth containing 1 mM isopropyl-β-d-thiogalactopyranoside at 37°C after 6 hrs (0.8 g/L of culture medium). The His-tag affinity chromatography followed by anti-His western blot analysis confirmed the purification of the mature His-tagged protein and the same-sized band was detected by the tuberculosis patient’s serum. Conclusion: The recombinant mature RpfE from M. tuberculosis lays the basis for immunogenicity studies as a vaccine candidate against tuberculosis.
INTRODUCTION
It is estimated that approximately one-quarter of the World’s population in 2014 were latently infected with Mycobacterium tuberculosis1. Reactivation of this clinically latent infection is responsible for a large proportion of active tuberculosis cases each year, with an estimated 1.4 million deaths in 20192.
The only licensed vaccine against tuberculosis is Mycobacterium bovis Bacillus Calmette-Guerin (BCG). However, BCG vaccination provides limited efficacy in preventing the establishment of latent infection or its subsequent reactivation3. This deficiency may be attributed to BCG’s inability to elicit sufficient immune responses against antigens associated with the latent and reactivation phases of the infection. Thus, incorporating candidate latency or reactivation antigens of M. tuberculosis into the development of new vaccines holds the potential to enhance the efficacy of anti-tuberculosis vaccination2.
Impairment of the immune responses in properly controlling mycobacterial growth in latently infected individuals is thought to cause tuberculosis reactivation. A family of five proteins in M. tuberculosis encoded by the rpf genes (rpfA-E) is able to induce resuscitation of the dormant bacilli4. Among the five mycobacterial Rpfs, RpfE is the smallest in size with 172 amino acids5, and is the focus of this study.
The RpfE is predicted to contain multiple B-cell epitopes, and numerous human histocompatibility leukocyte antigen (HLA) class I and HLA class II T-cell epitopes6-11. It is possibly antigen and non-allergen and is predicted to have a signal peptide cleavage site after residue 2812-14. It is predicted to be a secretory protein, most probably localized outside the cell, and has none or one transmembrane helix as shown by Hidden Markov Models5,15. This information from bioinformatics databases suggests RpfE is a potentially pertinent vaccine candidate, and worthy of production for antigenicity and immunogenicity studies.
Expression of recombinant proteins in Escherichia coli stands as a preferred, cost-effective, and straightforward approach for large-scale protein production. Thorough optimization of the expression process is crucial to minimize the efforts in downstream purification steps. These optimization procedures primarily focus on decreasing culture volume and maximizing the yield of soluble target proteins in their biological folding16.
The present study aimed to clone and optimally express full-length and mature RpfE proteins in E. coli towards obtaining purified recombinant protein, which can be used for further M. tuberculosis-specific immunogenicity studies and as a future vaccine candidate.
MATERIALS AND METHODS
Study area: This study was conducted in 2015 and 2016 at the Department of Microbiology, Faculty of Biological Sciences and Technology, Shahid Beheshti University, Tehran, Iran.
Molecular cloning of rpfE: The rpfE sequence from M. tuberculosis strain H37Rv was obtained from the accession number NP_216966.1. The whole gene sequence was chemically synthesized (Shingene, China). Restriction sites of BamHI and HindIII were incorporated in the sequence to allow in-frame cloning of the PCR product into pET-23a (+) plasmid (Novagen, Madison, Wisconsin, USA). For the expression of the mature RpfE protein (without the signal peptide), pET-28a (+) plasmid (Novagen) was employed. Amplification of fragment 84-516 (rpfE84-516) was carried out using the synthesized construct as a template and the EmeraldAmp Max HotStart PCR Master Mix (Takara Bio, Japan) according to the manufacturer’s instructions. The forward primer (5'-ATCGCGAATTCATGGACGACGCGGGCTTGGACCCA-3') had an EcoRI site (underlined) and a 5' start codon (double underlined) incorporated. The Reverse primer (5'-GCCTTGGTCTAGAAAGCTTGCCGCGG-3') put a HindIII site (underlined) at the 3' end of the fragment. Both full-length rpfE and rpfE84-516 fragments were cloned without a stop codon, leading to the incorporation of the polyhistidine (His) tag of pET-23a and pET-28a in the C-terminus of the recombinant proteins. Purified PCR products for both fragments were digested with the corresponding restriction enzymes (NEB, Ipswich, MA), were ligated into the cut vectors using T4 DNA ligase (NEB), and were transformed into Escherichia coli TOP10 cells (Invitrogen, Paisley, UK) by chemical method. The recombinant plasmids were isolated from individual colonies and were sequenced to confirm the correctly cloned coding sequence. The plasmids were then transformed into E. coli BL21 (DE3) (Invitrogen), and cells were stored at -70°C in 20% glycerol17.
Small scale expression and solubility testing: Recombinant E. coli BL21 strains were cultured overnight at 37°C in Luria-Bertani (LB) broth (Sigma-Aldrich, Missouri, USA) supplemented with 100 μg/mL ampicillin (Sigma-Aldrich) for pET-23a or 50 μg/mL kanamycin (Sigma-Aldrich) for pET-28a. They were then diluted (1:25) into fresh culture media containing the appropriate antibiotics grown to mid-log phase (OD at 600 nm~0.5-0.7). They were induced by adding 0.4 mM isopropyl β-D-thiogalactoside (IPTG) (Sigma-Aldrich) for pET-23a and 1 mM for pET-28a. Cultures were grown for an additional 10 hrs. The cells were harvested every hour by centrifugation (6000 g, 5 min), and the proteins from the supernatants were precipitated using increasing concentrations of ammonium sulfate (10 to 90% w/v) (Sigma-Aldrich)18. The cell pellets from 1 mL culture samples and the precipitates from 5 mL culture supernatants were added with 50 μL 1X phosphate-buffered saline (PBS, pH 7.4) (Sigma-Aldrich), mixed with 50 μL 2X Laemmli Sample Buffer (Bio-Rad, California, USA) plus 5% (v/v) β-mercaptoethanol (Sigma-Aldrich). They were heated for 10 min at 100°C. The cell lysates were divided into two groups, half of which was saved, and the supernatant of the other half was harvested by centrifugation (13,000 g, 30 min). Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) was employed on the culture supernatant precipitates, supernatants of cell lysate and total cell lysates, using the Amersham ECL Gel box and the Amersham ECL Gel 8-16% (GE Healthcare, Freiburg, Germany) following the manufacturer’s recommendations.
Samples were loaded into duplicate SDS-PAGE gels before electrophoresis. One of the gels was stained with 0.2% Coomassie brilliant blue R-250 (Sigma-Aldrich) for 1 hr and then destained in 5% methanol 7.5% acetic acid (Sigma-Aldrich). The protein bands from the other gel were blotted onto Polyvinylidene Difluoride (PVDF) membranes (Millipore, Bedford, USA) in a Semi-Dry Transfer Cell (Bio-Rad Laboratories Inc., California, USA) at 90 V for 1 hr in 1X Tris-Glycine buffer with 10% methanol (Sigma-Aldrich). The membrane was blocked for 1 hr at room temperature in Tris-buffered saline (TBS) containing 0.05% Tween 20 and 5% non-fat dry milk (Sigma-Aldrich). Immunostaining was performed for 1 hr at room temperature using Penta-His Mouse IgG1 monoclonal antibody (Cat No. 34660, Thermo Fisher Scientific, Waltham, MA) (diluted at 1:2000 in the blocking buffer), followed by 1 hr incubation at room temperature with goat anti-mouse IgG horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology Inc., California, USA) (1:5000 dilutions in the blocking buffer). The membrane was revealed with Western Blue Stabilized Substrate for Alkaline Phosphatase (Promega, USA), and bands were visualized using ImageQuant™ LAS 500 (GE Healthcare Life Sciences, Piscataway, New Jersey, USA).
Optimization of mature RpfE expression: The induction conditions (temperature and time after induction, and IPTG concentrations) were shown in Table 1. The expression level at each induction condition was compared with the total protein extract of non-induced cultures in 12% SDS-PAGE gels followed by Coomassie blue staining, and the band areas and intensities corresponding to mature RpfE were analyzed optically by ImageQuant TL and according to the manufacturer instruction.
| Table 1: | Different concentrations used for the optimization of mature RpfE expression | |||
| Induction temperature (°C) | Induction time (hr) at 37°C | IPTG concentration (mM) |
| 37 | 1.5 | 0.25 |
| 35 | 3 | 0.5 |
| 30 | 4.5 | 1 |
| - | 6 | 1.25 |
| - | - | 1.5 |
Large-scale production of mature RpfE: Recombinant BL21 cells carrying pET28a/rpfE84-516 expressing the mature RpfE with His-tags at C- and N-terminus were grown in 200 mL LB-broth in 1 L baffled flasks at 37°C to an OD600 of 0.6, were induced with 1 mM IPTG and the cells were harvested after 6 hrs by centrifugation and stored at -20°C. Bacterial pellets were then thawed and disrupted by sonication (20 sec, 6 times) in a lysis buffer (20 mL/g cell pellet) composed of 20 mM NaH2PO4, 500 mM NaCl, and 6 M Urea (pH 8), followed by centrifugation (13000×g, 30 min, 4°C) to remove cell debris. The complete His-Tag Purification Column (Roche, Mannheim, Germany) was utilized for protein purification following the manufacturer’s guidelines. Elution of the protein was carried out using a column volume of 500 mM imidazole at pH 8, supplemented with a complete protease inhibitor (Sigma-Aldrich), and subsequently analyzed by SDS-PAGE. The protein fractions obtained through affinity purification were combined and subjected to dialysis against PBS (pH 7.4). Quantification was performed using the Bradford total protein assay kit (ZellBio GmbH, Ulm, Germany). Assessment for LPS contamination was conducted through the Limulus amoebocyte lysate (LAL) assay (Cambrex, USA).
Western blot analysis with tuberculosis patient serum: The dialyzed sample was resolved on a 12% SDS-PAGE gel and transferred onto a PVDF membrane. Western blotting was carried out as mentioned above. The membrane was incubated with serum from a consented tuberculosis-positive case (1:50 dilution in TBS) for 2 hrs at room temperature, and with HRP-conjugated rabbit anti-human IgG (Dako, Glostrup, Denmark) in a dilution of 1:5000 (in TBS) for 2 hrs at room temperature. The protein band was developed using diaminobenzidine (DAB) substrate (0.1% H2O2+10 mg DAB in 10 mL of TBS).
RESULTS AND DISCUSSION
Comparing precipitates from culture supernatants (Fig. 1-3a-b) with total cell lysate (Fig. 1-3c-d) revealed that the full-length recombinant RpfE protein was soluble and secreted into broth medium, resulting in a band of approximately 26 kDa in SDS-PAGE and western blots. Various alterations made to the culture conditions included incubation at 37°C with 0.1 and 0.2 mM IPTG for 8 to 11 hrs (Fig. 1), at 37°C with 0.3 and 0.4 mM IPTG for 8 to 11 hrs (Fig. 2), and at 30 C with 0.1 to 0.4 mM IPTG for 13 to 16 hrs (Fig. 3). These alterations did not yield any difference in the localization of the full-length protein. The best ammonium sulfate concentration for protein precipitation was 70% (Fig. 4).
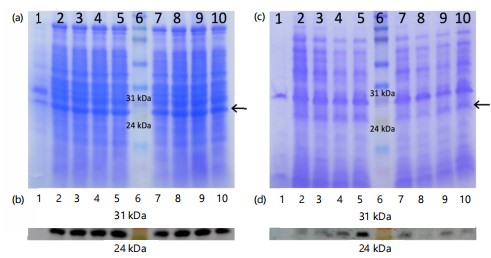
|
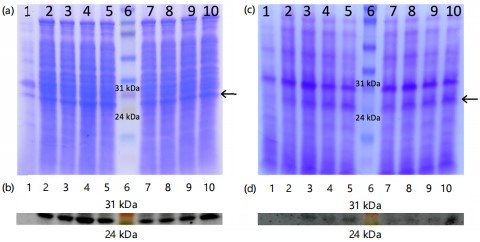
|
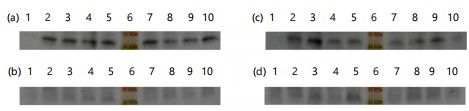
|
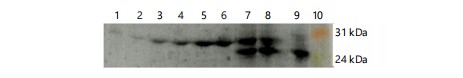
|
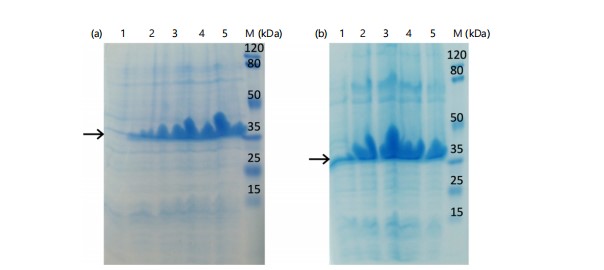
The recombinant mature RpfE protein encoded by pET-28a/rpfE84-516 expression vector was soluble, accumulated in the cytoplasm of E. coli BL21 and multimerized as a band of ~35 kDa. Optimization of induction conditions gave the highest expression of mature RpfE at 37°C after 6 hrs, with 1 mM IPTG in
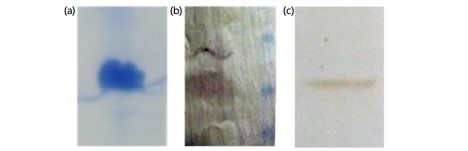
LB broth with a yield of 0.8 g/L of culture medium (Fig. 5). The mature RpfE protein was successfully purified (Fig. 6a) and was detected with anti-His antibody (Fig. 6b) and with the serum from a tuberculosis patient (Fig. 6c). The endotoxin content of the recombinant protein was <50 IU/mg.
Comprehensive data mining and bioinformatic analysis have identified RpfE protein as the second most promising candidate among 189 putative vaccine candidates chosen from the entire genome of M. tuberculosis19. Likewise, RpfE was selected as a pertinent vaccine candidate in another study20. These bioinformatic predictions have been endorsed by an in vitro study showing that RpfE can induce dendritic cell (DC) maturation21. These mature DC promoted naïve CD4+ T-cell differentiation towards T helper (Th)1 and Th17 cells which have been shown to play an important role in establishing protective immunity against tuberculosis22. Furthermore, a study with RpfE immunized mice presented a significant level of protection against high-dose intravenous challenge with M. tuberculosis strain H37Rv regarding mycobacterial loads in lungs and spleens and post-infection survival periods23. Altogether, RpfE exhibits a promising potential to be incorporated into new tuberculosis vaccines. Accordingly, it worthwhile to clone and optimize the expression to produce purified recombinant RpfE for future vaccination studies.
|
|
Full-length rpfE (encoding 172 amino acids, including the signal peptide sequence) was firstly cloned into pET-23a and expressed in E. coli BL21. The resulting 26 kDa recombinant protein was soluble and was secreted into the broth medium. The E. coli apparatus cleaved the signal sequence of the full-length RpfE protein. Although RpfE is a mycobacterial protein, a signal peptide cleavage site between the residues 28 and 29 is also predicted in E. coli, which can explain the secretion of the recombinant protein into the medium14. Full-length rpfE was previously cloned in E. coli DH5α by Ying and colleagues, expressing a 22 kDa protein similar to our yield. Still, unlike current study results, their highly expressed recombinant RpfE was accumulated inside the cells at approximately 16% of total cellular protein24. In the present study, that high expression of full-length RpfE in E. coli was not achieved. Therefore, the gene lacking the coding sequence for signal peptide (nucleotides 84 to 516, encoding 144 amino acids) was cloned into pET-28a. The mature protein was expressed and accumulated in soluble form in the cytoplasm of E. coli. The resulting small recombinant protein tended to multimerize and produced a band of ~35 kDa in SDS-PAGE gel. This is probably due to two cysteine residues in its sequence5 disabling the β-mercaptoethanol in the sample buffer to break multiple disulfide bonds. Comparison between the bands of full-length and mature RpfE indicated that the presence of signal peptide reduced the expression of the protein (more than a 5-fold decrease). It has been suggested that signal peptide interferes with the production of large quantities of functional proteins in E. coli, which could reduce or even hinder the expression of the recombinant proteins25. Altogether, the higher levels of soluble mature RpfE compared to full-length RpfE convinced us to use the protein without its signal sequence for further optimization experiments.
Recombinant protein expression must be optimized thoughtfully to minimize efforts in subsequent large-scale purification steps16,26. Firstly, different induction temperatures were tested and an obvious decline in the protein expression was observed by decreasing the temperature from 37 to 35°C (data not shown) and 30°C (Fig. 3; shown is the data for full-length RpfE). Therefore, the induction time and IPTG concentration for mature RpfE were optimized at 37°C (Fig. 5). The highest yield for mature RpfE was achieved by adding 1 mM IPTG for 6 hrs at 37°C in LB-broth. Under these conditions, levels of mature RpfE expression in E. coli BL21 was ~0.8 g/L of culture medium, which is within the range of the highest yields of soluble recombinant proteins described in the literature27. Choi and colleagues also reported cloning of RpfE into pET-22b(+) in E. coli without its signal sequence21. They showed N-terminal His-tagged RpfE with an approximate molecular weight of 14 kDa which was localized primarily in inclusion bodies21. Interestingly, the same induction conditions were used to express mature RpfE in E. coli BL21 as that optimized in current study.
Purification of mature RpfE was done in a denaturing condition by the addition of 6 M urea to the lysis buffer. Similarly, both previously mentioned studies on recombinant RpfE used denaturing conditions and urea for protein purification21,24. At the same time, mature RpfE was successfully purified and eluted from the His-tag column in its native form as it was detected by serum from a tuberculosis patient in western blot analysis.
The purified RpfE protein produced in this study holds promise as a potential vaccine candidate. This application was facilitated by selecting the optimal form of the protein for expression in E. coli and optimizing its expression. Our study presents a pipeline for future research aiming to produce mycobacterial proteins in E. coli. While the purified mature RpfE was successfully detected by serum from a tuberculosis patient in western blot analysis, further investigation is required to determine if the untreated protein can stimulate T cells as an antigen and to assess its immunogenic potential upon administration in animal models. This area remains a focus for subsequent research by our team.
CONCLUSION
This study describes that the purified mature His-tagged RpfE recombinant protein from M. tuberculosis was efficiently expressed in E. coli. This purified recombinant protein might be a good candidate for vaccine studies against tuberculosis. Further studies are encouraged to evaluate the stability and profile of immune responses after antigenicity studies and after immunizing mice with this RpfE protein.
SIGNIFICANCE STATEMENT
Mycobacterium tuberculosis RpfE is a possibly secretory protein and that presents a challenge in high expression of this protein in its biological form in E. coli. This study overcame this challenge by cloning the full-length gene compared to a gene fragment lacking the signal peptide (encoding mature RpfE). Subsequently, a systemic optimization of the expression conditions for mature RpfE enabled achieving a high yield of the soluble protein. The protein was successfully purified, and its native folding was confirmed by its interaction with serum from a consented tuberculosis patient. The purified protein is a potential candidate for vaccination studies against tuberculosis.
REFERENCES
- Houben, R.M.G.J. and P.J. Dodd, 2016. The global burden of latent tuberculosis infection: A re-estimation using mathematical modelling. PLoS Med., 13.
- Chakaya, J., M. Khan, F. Ntoumi, E. Aklillu and R. Fatima et al., 2021. Global tuberculosis report 2020-Reflections on the global TB burden, treatment and prevention efforts. Int. J. Infect. Dis., 113: S7-S12.
- WHO, 2018. BCG vaccine: WHO position paper, February 2018-Recommendations. Vaccine, 36: 3408-3410.
- Mukamolova, G.V., O.A. Turapov, D.I. Young, A.S. Kaprelyants, D.B. Kell and M. Young, 2002. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol. Microbiol., 46: 623-635.
- Lew, J.M., A. Kapopoulou, L.M. Jones and S.T. Cole, 2011. TubercuList-10 years after. Tuberculosis, 91: 1-7.
- Larsen, J.E.P., O. Lund and M. Nielsen, 2006. Improved method for predicting linear B-cell epitopes. Immunome Res., 2.
- Saha, S. and G.P.S. Raghava, 2006. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins: Struct. Funct. Bioinf., 65: 40-48.
- Larsen, M.V., C. Lundegaard, K. Lamberth, S. Buus, O. Lund and M. Nielsen, 2007. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinf., 8.
- Kobayashi, H., M. Wood, Y. Song, E. Appella and E. Celis, 2000. Defining promiscuous MHC class II helper T-cell epitopes for the HER2/neu tumor antigen. Cancer Res., 60: 5228-5236.
- Singh, H. and G.P.S. Raghava, 2001. ProPred: Prediction of HLA-DR binding sites. Bioinformatics, 17: 1236-1237.
- Rammensee, H.G., J. Bachmann, N.P.N. Emmerich, O.A. Bachor and S. Stevanović, 1999. SYFPEITHI: Database for MHC ligands and peptide motifs. Immunogenetics, 50: 213-219.
- Doytchinova, I.A. and D.R. Flower, 2007. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinf., 8.
- Saha, S. and G.P.S. Raghava, 2006. AlgPred: Prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res., 34: W202-W209.
- Petersen, T.N., S. Brunak, G. von Heijne and H. Nielsen, 2011. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods, 8: 785-786.
- Rashid, M., S. Saha and G.P.S. Raghava, 2007. Support vector machine-based method for predicting subcellular localization of mycobacterial proteins using evolutionary information and motifs. BMC Bioinf., 8.
- Sørensen, H.P. and K.K. Mortensen, 2005. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb. Cell Fact., 4.
- Green, M.R. and J. Sambrook, 2012. Molecular Cloning: A Laboratory Manual. 4rd Edn., Cold Spring Harbor Laboratory Press, Long Island, New York, ISBN: 978-1-936113-42-2, Pages: 2028.
- Duong-Ly, K.C. and S.B. Gabelli, 2014. Salting Out of Proteins Using Ammonium Sulfate Precipitation. In: Methods in Enzymology, Neil, E. and G. Marsh (Eds.), Elsevier Inc., Amsterdam, Netherlands, ISBN: 9780124201194, pp: 85-94.
- Zvi, A., N. Ariel, J. Fulkerson, J.C. Sadoff and A. Shafferman, 2008. Whole genome identification of Mycobacterium tuberculosis vaccine candidates by comprehensive data mining and bioinformatic analyses. BMC Med. Genomics, 1.
- Thi, L.T.N., M.E. Sarmiento, R. Calero, F. Camacho and F. Reyes et al., 2014. Immunoinformatics study on highly expressed Mycobacterium tuberculosis genes during infection. Tuberculosis, 94: 475-481.
- Choi, H.G., W.S. Kim, Y.W. Back, H. Kim and K.W. Kwon et al., 2015. Mycobacterium tuberculosis RpfE promotes simultaneous Th1-and Th17-type T-cell immunity via TLR4-dependent maturation of dendritic cells. Eur. J. Immunol., 45: 1957-1971.
- Griffiths, K.L., A.A. Pathan, A.M. Minassian, C.R. Sander and N.E.R. Beveridge et al., 2011. Th1/Th17 cell induction and corresponding reduction in ATP consumption following vaccination with the novel Mycobacterium tuberculosis vaccine MVA85A. PLoS ONE, 6.
- Yeremeev, V.V., T.K. Kondratieva, E.I. Rubakova, S.N. Petrovskaya and K.A. Kazarian et al., 2003. Proteins of the Rpf family: Immune cell reactivity and vaccination efficacy against tuberculosis in mice. Infect. Immun., 71: 4789-4794.
- Xue, Y., Y. Bai, X. Gao, H. Jiang, L. Wang, H. Gao and Z. Xu, 2012. Expression, purification and characterization of Mycobacterium tuberculosis RpfE protein. J. Biomed. Res., 26: 17-23.
- Larentis, A.L., A.P.C. Argondizzo, G. dos Santos Esteves, E. Jessouron, R. Galler and M.A. Medeiros, 2011. Cloning and optimization of induction conditions for mature PsaA (pneumococcal surface adhesin A) expression in Escherichia coli and recombinant protein stability during long-term storage. Protein Expression Purif., 78: 38-47.
- Ashayeri-Panah, M., F. Eftekhar, B. Kazemi and J. Joseph, 2017. Cloning, optimization of induction conditions and purification of Mycobacterium tuberculosis Rv1733c protein expressed in Escherichia coli. Iran. J. Microbiol., 9: 64-73.
- Choi, J.H., K.C. Keum and S.Y. Lee, 2006. Production of recombinant proteins by high cell density culture of Escherichia coli. Chem. Eng. Sci., 61: 876-885.
How to Cite this paper?
APA-7 Style
Ashayeripanah,
M., Saubi,
N., Joseph,
J., Eftekhar,
F., Kazemi,
B. (2025). Cloning, Optimized Expression and Purification of Mycobacterium tuberculosis RpfE Protein in Escherichia coli. Asian Science Bulletin, 3(2), 119-127. https://doi.org/10.3923/asb.2025.119.127
ACS Style
Ashayeripanah,
M.; Saubi,
N.; Joseph,
J.; Eftekhar,
F.; Kazemi,
B. Cloning, Optimized Expression and Purification of Mycobacterium tuberculosis RpfE Protein in Escherichia coli. Asian Sci. Bul 2025, 3, 119-127. https://doi.org/10.3923/asb.2025.119.127
AMA Style
Ashayeripanah
M, Saubi
N, Joseph
J, Eftekhar
F, Kazemi
B. Cloning, Optimized Expression and Purification of Mycobacterium tuberculosis RpfE Protein in Escherichia coli. Asian Science Bulletin. 2025; 3(2): 119-127. https://doi.org/10.3923/asb.2025.119.127
Chicago/Turabian Style
Ashayeripanah, Mitra, Narcis Saubi, Joan Joseph, Fereshteh Eftekhar, and Bahram Kazemi.
2025. "Cloning, Optimized Expression and Purification of Mycobacterium tuberculosis RpfE Protein in Escherichia coli" Asian Science Bulletin 3, no. 2: 119-127. https://doi.org/10.3923/asb.2025.119.127

This work is licensed under a Creative Commons Attribution 4.0 International License.




